EPIGENETIC CONTROL OF T‐CELL ACTIVATION
Epigenetic control of gene expression regulates T‐cell activation and differentiation
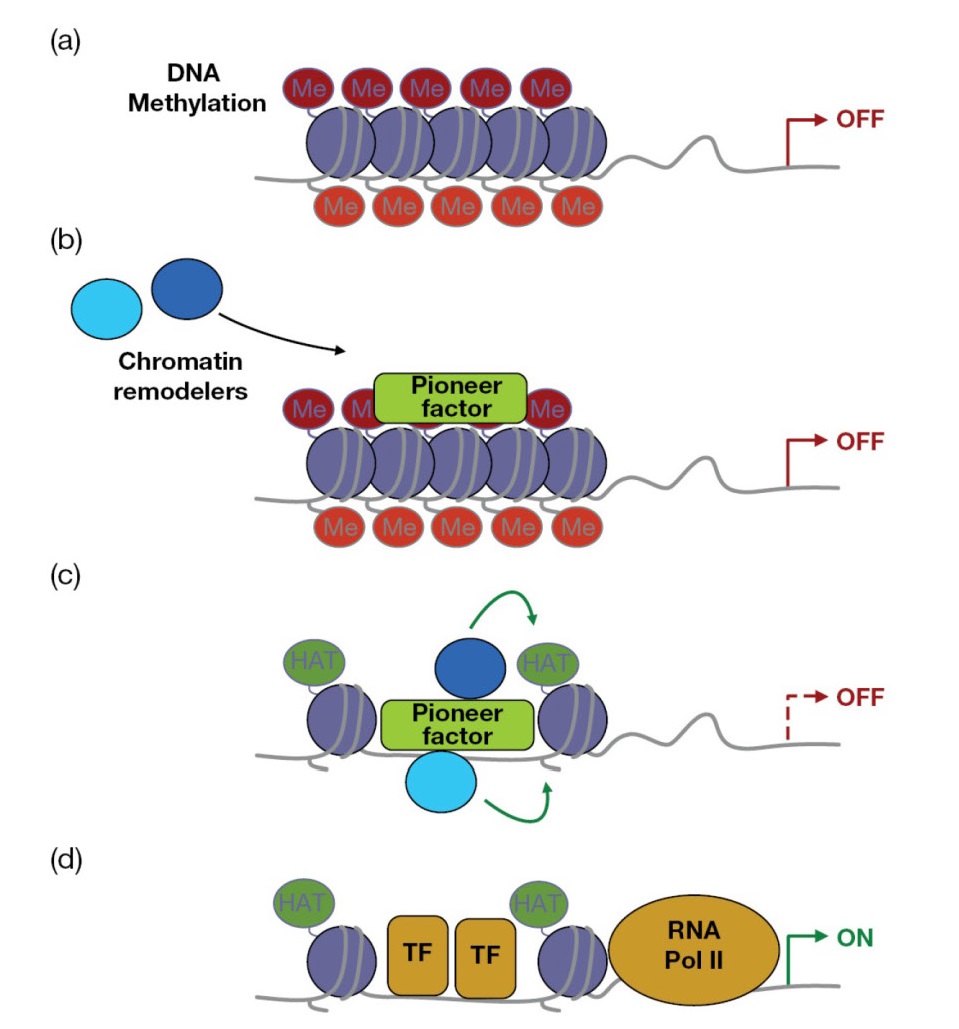
Activation and differentiation of T‐cells into the correct effector subsets is fundamental to generating an immune response capable of fighting a specific infection. Accordingly, the genes controlling T‐cell activation and differentiation are tightly controlled. Nuclear DNA is normally wrapped around proteins called histones, which act as spools around which DNA winds, allowing the cell to compact and order a large amount of genetic information into the relatively small confines of the nucleus. Importantly, histones act as guardians of genetic information by shielding genes from activating transcription factors and as such, histone modification introduces a important layer of regulation of gene expression. For example, posttranslational modifications of histones at specific amino acids may directly change the conformation of histone at that site and effectively loosen or tighten its grip on DNA, thereby making it more or less accessible for transcription factor binding and gene activation. This can also occur indirectly, where histone modification creates a binding site for chromatin‐modifying factors, which can then change the structure of chromatin to activate or repress gene transcription at a particular locus. ChIP‐sequencing (Chip‐Seq) is an experimental technique that combines chromatin immunoprecipitation with large‐scale DNA sequencing to detect binding sites between proteins and DNA on a genome‐wide scale. This technology has uncovered many important histone modifications, including trimethylation of histone H3 at lysine 4 (H3K4me3), which promotes an active chromatin arrangement at particular genes, and H3K27me3, which may tighten chromatin and repress gene transcription. In addition, direct methylation of DNA at CpG sites may render genes less transcriptiona ly active and this can play an important role in gene regulation.
Epigenetic factors controlling T‐cell
polarization
Binding of DNA
methyltransferases promotes the transfer of a methyl group to DNA CpG islands,
which can act as a platform for binding of methyl‐CpG‐binding
domain proteins (MBDs). These proteins play an important role in gene
regulation by recruiting chromatin remodeling factors that can compact the
chromatin around a particular gene, thereby suppressing gene induction.
Repressive regulation at the I14 gene locus appears to play an important
role in shaping the outcome of differentiation. Binding of the DNA
methyltransferase Dnmt‐1 to the I14 locus
recruits MBD2 to compact chromatin around the I14 gene, blocking
transcription factor access. Accordingly, genetic deletion of either of these
factors leads to aberrant, GATA3‐ independent I14 expression in non‐Th2 subsets. The I14 locus is further
repressed by H3K27me3 in Th1 cells, while Th2 cells have activating H3K4Me3 at
the I14 gene, thus promoting I14 transcription and the Th2
phenotype. Supporting Th1 polarization, activating H3K4Me3 modifications have
been found at the Ifng locus in Th1 cells, which promote binding of the
Th1 master regulator T‐bet, with associated Ifnγ gene transcription.
Conversely, transcription of Ifnγ in Th2 cells is repressed
through DNA methylation, and trimethylation of histone H3K27me3, thus
preferentially skewing towards Th2 differentiation in the presence of Th2
cytokines. Epigenetic control also extends to Th17 and Treg cells, which
display activating H3K4Me3 at the Rorγt and Foxp3 genes
respectively, together with repressive H3K27me3 marks at both gene loci in non‐expressing Th types. In this
regard, deacetylation of histones and subsequent repression of gene activity at
the Foxp3 locus seems to play an important role in regulating Treg
numbers, as mice deficient in the Foxp3 deacetylase sirtu in 1 have increased numbers of Tregs and
enhanced immunosuppression.
As might be expected,
transcription of the master regulators of Th1 and Th2 programming, T‐bet and GATA3 also appear to
be regulated by strong activating H3K4Me3 marks in each respective subtype,
with the absence of inactivating H3K27me3 at these loci. Interestingly,
together with inactivating H3K27me3, the T‐bet and Gata3 loci are
also decorated with a small amount of activating
H3K4Me3 in non‐expressing subtypes.
Decoration of histones with both activating and inactivating marks at the same
gene is indicative of bivalent genes and indicates that these master regulators
may be “poised” to undergo expression in non‐expressing lineages when the occasion arises. This creates the intriguing
possibility that activating and inhibitory histone modifications act as a
switch to enhance the degree of plasticity of differentiation that has been
observed between Th subtypes.
Enhancer elements are
noncoding regions of genes that recruit transcription factors to turn on gene
expression. These important determinants of cell type specificity have proved
elusive to experimentally locate but recent developments in chromatin signature identification have facilitated
genome‐wide mapping of gene enhancers. H3K4Me1 marks have been
identified as an activating enhancer signature, as has the binding of
acetyltransferase p300, both of which open up the enhancer region for
transcription factor binding. A key question in T‐cell biology has been how signals from the external
and intracellular environment modulate the enhancer landscape, and thus
activation, of master genes. Recent work has elegantly addressed this issue by
using p300 and H3K4Me1 as probes in a genome wide screen of enhancer regulators during CD8+ T‐cell differentiation. While
STAT‐induced T‐bet and GATA3 have been
assumed to activate key Th1 and Th2 genes in a linear fashion, it now appears
that STATs have a greater role to play than simply activating these master
regulators. STATs have been found to promote binding of p300 to key Th1 and Th2
enhancers, and this was largely required to make the enhancer accessible for
subsequent binding and activation by T‐bet or GATA3, with H3K4Me1 playing a lesser role. Importantly, induced
expression of T‐bet or GATA3 was
insufficient to activate expression of Th1 or Th2 genes in STAT‐deficient cells, suggesting
that in addition to activating key transcription factors, STAT proteins may act
as “pioneer” factors that open up the enhancer landscape r quired for gene
expression and differentiation (Figure 7.15).