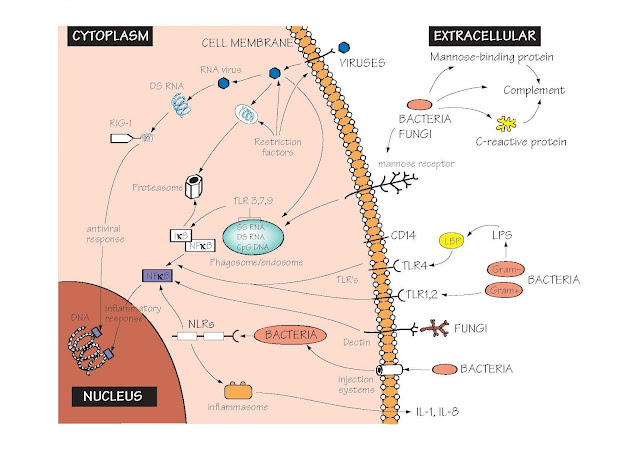
Receptors Of The Innate Immune System
The ability to sense the presence of
microorganisms that could cause potentially
dangerous infections is a widespread property of cells, tissues and body fluids
of all multicellular organisms. This process is called innate immune
recognition. This recognition process is the first crucial step triggering
the complex sequence of events by which the body protects itself against
infection. However, it is only since the 1980s that most of the molecules
(receptors) responsible for this recognition process have been identified, and
new examples of such innate receptors are still being found. The receptors
usually recognize components of microorganisms that are not found on cells of
the host, e.g. components of
bacterial cell wall, bacterial flagella or viral nucleic acids. These target
molecules have been named pathogen-associated molecular patterns (PAMPS), and the receptors that recognize them
pattern recognition receptors (PRRs). Engagement of PRRs by PAMPs results in
activation of intracellular signalling pathways, resulting in alteration in
gene transcription in the nucleus (left part of figure) and ultimately a whole
variety of different cellular responses, broadly termed inflammation (illustrated
in Fig. 7). Some innate immune receptors are also triggered by damage to cells
that arises in the absence of any infection, giving rise to the term
damage-associated molecular patterns (DAMPs). The activation of innate immunity
is an essential prerequisite for activation for most adaptive immune responses.
The major families of PRRs, the structures they recognize and their location within the cell are shown.
Leucine-rich repeats (LRR) A ubiquitous protein structural motif, forming a ‘horseshoe’-shaped fold, with an
exposed hydrophilic surface and a tightly packed internal hydrophobic core. It
is so named because it contains unusually large numbers of the hydrophobic
amino acid leucine. LRRs are frequent components of PRRs, where they are
thought to mediate the interaction between the receptor and the target
structure on the microorganism. Families of proteins containing LRRs may also
serve primitive antibody-like functions in several types of invertebrates (see
Fig. 46).
Toll-like receptors (TLR) Toll-like receptors are so named because of
their homology to a gene named Toll (from the German word for ‘amazing’
or ‘mad’!) first identified in Drosphila. TLRs were the first PRRs to be
discovered, and have come to represent the archetype of innate immune
recognition receptors. Humans have 10 TLRs, each with an LRR domain involved in
recognition of microbial components, and an intracytoplasmic TIR domain
involved in signalling into the cell. TLRs associate with a variety of adaptor
molecules that help to convert recognition of microbes into a signal, which
activates specific gene transcription within the cell.
RIG-1 Many viruses carry their genetic information in
the form of RNA, rather than DNA as do all eukaryotes. RIG-1 is an example of a
family of molecules that recognize RNA viruses such as influenza,
picornaviruses (common cold) and Japanese encephalitis virus, and then switch
on the production of interferons and other antiviral proteins (see Fig.
23).
Cell surface Innate recognition receptors at the cell
surface recognize extracellular microorganisms. The best studied example is
TLR4, which together with accessory molecules MD2 and CD14, recognizes
lipopolysaccharide (LPS), the principal component of Gram-negative bacterial
walls. TLR4 is distributed on many cell types, but is especially important on
macrophages (see Figs 7 and 8). Excessive activation of macrophages is thought
to be a major factor in sepsis and endotoxic shock, which leads
to oedema and low blood pressure, and can be fatal.
Cytoplasm Many microorganisms can efficiently cross the
cellular membrane and colonize the cytoplasm. Viruses are the best known examples
of cytoplasmic pathogens. However, many bacteria can also either cross the
membrane into the cytoplasm (e.g. Salmonella) or can inject toxins and
other bacterial components into the cytoplasms. Intracytoplasmic bacterial
components are recognized by the NOD- like receptors.
NOD-like receptors These are a large family of cytoplasmic
proteins that contain leucine-rich repeats, which bind to bacterial
components. NOD1 and NOD2 recognize fragments of bacterial cell wall prote-
oglycans, and are found at particularly high amounts in the epithelial cells
that line the gut. Mutations in NOD2 have been found to increase the likelihood
of developing Crohn’s disease, a chronic inflammatory gut disease, perhaps
because of a deficient response to bacteria in the gut. Some NOD-like receptors
activate the transcription factor NFκB. Others activate the inflammasome.
The inflammasome This is a multimolecular complex that is assembled
in response to triggering of some NOD-like receptors, and leads to the
secretion of active forms of the inflammation-promoting cytokines IL-1 and IL-18
(see Fig. 23). Persistent activation of the inflammasome by crystals of uric
acid is thought to cause many of the symptoms of gout. In some cases,
activation of the inflammasome results in the rapid death of the host cell by a
special process known as pyroptosis.
Restriction factors A collection of proteins that inhibit the
ability of viruses to replicate. Trim5α binds retroviruses and carries them to
the proteasome, an intracellular organelle that destroys them. Tetherin,
as its name suggests, binds to some viruses as they bud off from the cell
surface, limiting the ability of the virus to spread. New restriction factors
are continually being discovered.
The endosome/phagosome Many microorganisms are taken up by endocytosis
or phagocytosis by macrophages (see Fig. 9). Several TLRs sense microorganisms
within these compartments. TLR9 recognizes a type of DNA found predominantly in
bacteria and viruses, but rare in eukaryotes (CpG DNA). TLR3 recognizes
double-stranded RNA, found in many viruses. TLR7 recognizes single-stranded
RNA, which is found in many RNA viruses. Although single-stranded RNA is also a
ubiquitous component of eukaryotic cells, it is unstable and cannot survive in
the extracellular environment. It therefore seldom enters the
endosomal/phagocytic system.
CRP C-reactive protein (MW 130 000), a pentameric
globulin (or ‘pentraxin’) made in the liver which appears in the serum within
hours of tissue damage or infection, and whose ancestry goes back to the
invertebrates. It binds to phosphorylcholine, which is found on the surface of
many bacteria, fixes complement and promotes phagocytosis (see Fig. 6).
Mannose-binding lectin (MBL) A serum protein that binds the sugar mannose,
which is often found in large amounts on bacterial or fungal surfaces, but is
usually not exposed on mammalian cells. Binding of MBP to microbial surfaces
then activates complement (see Fig. 6).
NFκB NFκB is a key transcription factor regulating
the inflammatory response. Normally, it is kept inactive in the cytoplasm by
binding to the inhibitor IκB. However, activation of many PRRs (see
figure) results in destruction of IκB by the proteasome, and NFκB then
moves into the nucleus where it switches on many components of the
antibacterial, antiviral and inflammatory response.
Proteasome A cytoplasmic organelle whose major function is
to break down proteins and recycle their constituent amino acids within the
cell. It also has a key role in producing the peptides recognized by the T
lymphocyte (see Fig. 18).
Dectin-1 and the mannose
receptor These are just two
members of an enormous family of sugar-binding proteins known as C-type
lectins. They have an important role in binding to fungal and bacterial cell
walls, activating phagocytosis and inflammation (see Figs 8 and 9). walls, activating phagocytosis and inflammation
(see Figs 8 and 9).